Changes and New Features¶
This section describess the new and updated functionality present in version 3.4.0 of ProtoMS.
GCAP Simulations¶
ProtoMS is now able to perform grand canonical alchemical perturbation (GCAP) simulations, where ligand perturbations can be performed with GCMC water sampling. These can be set up using protoms.py -s gcap_single or protoms.py -s gcap_dual, for single and dual topology calculations, respectively. These can either be performed at a range of 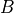 values, known as surface-GCAP (two-dimensional simulation in
values, known as surface-GCAP (two-dimensional simulation in 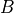 and
and  ) or at the equilibrium
) or at the equilibrium 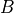 value, which is single-GCAP. These new simulation methods come with a new analysis tool
value, which is single-GCAP. These new simulation methods come with a new analysis tool calc_gcap_surface.py which is able to calculate the free energy from the two-dimensional GCAP surface.
Grand Canonical Integration¶
More automation has been introduced to calc_gci.py to calculate the appropriate number of steps for the titration fit. The water occupancy at the equilibrium 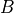 value is also returned. Considerable reduction in the noise of gcmc calculations has been achieved thanks to replica exchange allowing the removal of the absolute and pseudo-huber error functions. Error reporting for model ensembles has also been removed.
value is also returned. Considerable reduction in the noise of gcmc calculations has been achieved thanks to replica exchange allowing the removal of the absolute and pseudo-huber error functions. Error reporting for model ensembles has also been removed.
Free Energy Calculations¶
Updates have been made to the analysis of free energy calculations. calc_ti_decomposed.py allows the different contributions to the free energy changes to be extracted. This is useful to understand where changes in relative free energies are arising from. calc_dg_cycle.py allows the cycle closure and free energy changes of various legs to be determined for a set of free energy perturbations.
Additionally, a ‘partial’ implementation of dual topology calculations is now available. If the change between two ligands in a relative free energy calculation is too large for single topology, but dual topology would not be appropriate, it is now possible to apply softcore potentials to only the changing groups, whilst leaving the common structure unchanged, reducing noise in the calculated free energy.
protomslib¶
The underlying class structures and many of the fundamental functions of the Python layer of ProtoMS have now been restructured into an importable module, protomslib. This also allows for support of Python  3.5 (retaining support for Python 2.7) with the setup and analysis scripts.
3.5 (retaining support for Python 2.7) with the setup and analysis scripts.
Restarting Simulations¶
The writing of restart files in ProtoMS has now been significantly improved. Simulations which were terminated early or did not complete. When restarting from an aborted simulation, ProtoMS will now perform only the necessary Monte Carlo steps to complete the simulation, instead of just using the structural data as the start of a new run.
Water Clustering¶
The clustering of GCMC waters with calc_clusters.py has been changed to prevent two waters observed in the same frame from being placed in the same cluster. This previously allowed for cluster occupancies of greater than 100%, and has now been corrected. The clustering should also now be less sensitive to the distance cutoff used.
