
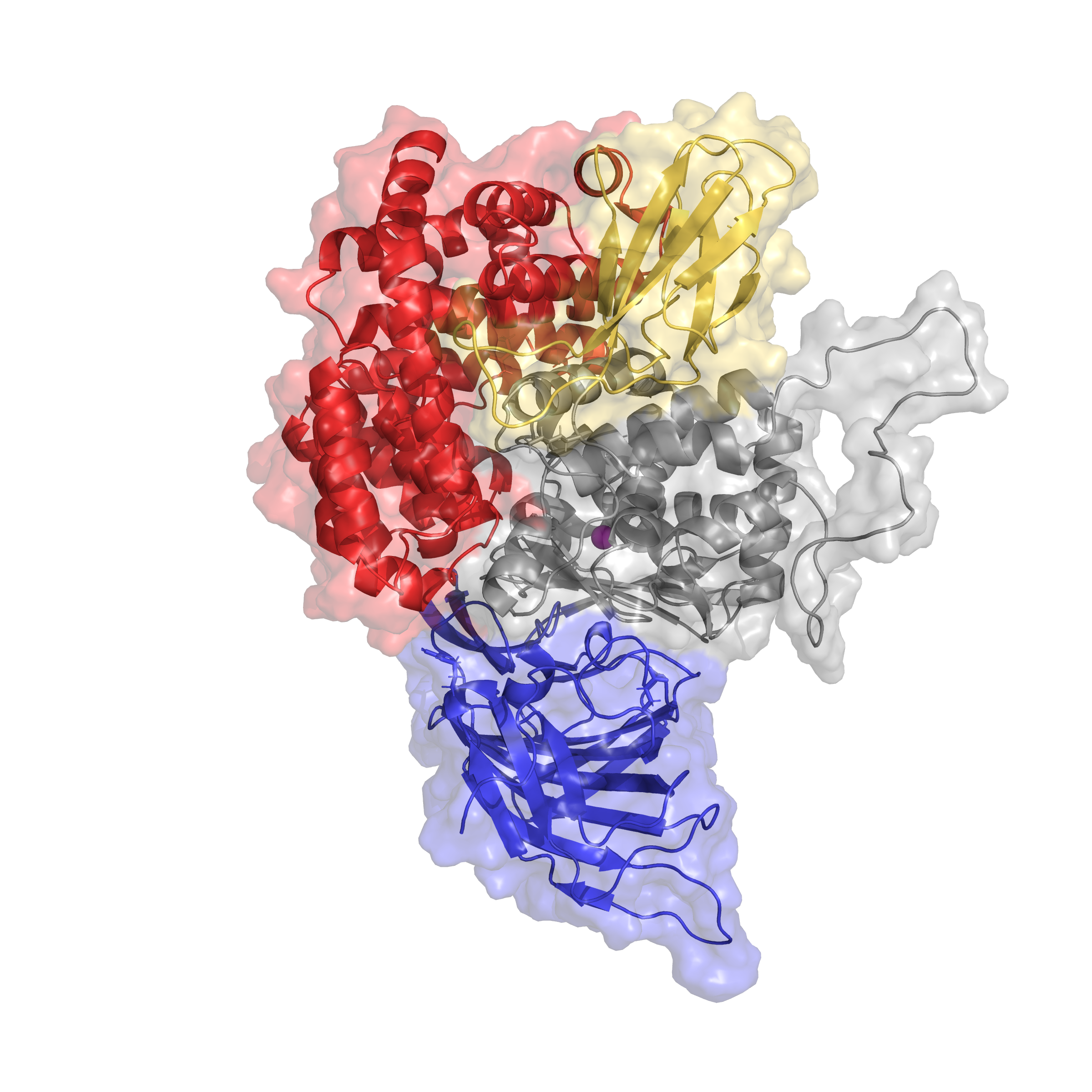
My work in the Essex group has involved small molecule parametrisation and docking, model building from crystal structures, and molecular dynamics simulations, which I use to investigate ligand binding in Endoplasmic Aminopeptidase 1. ERAP1 is a multifunctional enzyme and has complex substrate specificity including peptide substrate length and amino acid sequence.I also investigate the polymorphic nature of ERAP1 at the molecular level. ERAP1 variants vary in substrate preference and trimming rates, and are associated with autoimmune disease, hypertension, and cancer prognosis. The relationship between protein structure and trimming phenotype in ERAP1-associated disease is not fully understood and could provide valuable insight for rational inhibitor design and treatment.Greater understanding of this enzyme, its catalytic mechanism and ligand binding has improved design of molecular and cell-based experiments in the lab including site-directed mutagenesis and enzymatic assays. These in vitro experiments are carried out with the Elliott and James lab in the Cancer Sciences department which specialises in head and neck cancer-associated ERAP1 variants, and peptide processing and presentation on MHCI molecules.