Tools¶
In the $PROTOMSHOME/tools folder we have collect a range of useful scripts to setup and analyse ProtoMS simulations. Many of them are used by the protoms.py setup script. In this page we have collected the documentation for these tools with the user as a focus. Developers might be interested in looking at the Python code manual in the .doc folder.
ambertools.py¶
Program to run antechamber and parmchk for a series of PDB-files
usage: ambertools.py [-h] [-f FILES [FILES ...]] [-n NAME]
[-c CHARGE [CHARGE ...]]
Named Arguments¶
| -f, --files | the name of the PDB-files |
| -n, --name | the name of the solute Default: “UNK” |
| -c, --charge | the net charge of each PDB-file |
Examples:
ambertools.py -f benzene.pdb
ambertools.py -f benzene.pdb -n BNZ
ambertools.py -f benzenamide.pdb -c 0
ambertools.py -f benzene.pdb toluene.pdb -n BNZ TOL
Description:
This tool encapsulate the program antechamber and parmchk from the AmberTools suite of programs.
It will produce an Amber prepi-file, containing the z-matrix and atom types of the given solutes, parametrized with the general Amber force field and AM1-BCC charges. It will also produce an Amber frcmod-file with additional parameters not found in the GAFF definition. These files be named after the input pdbfile, replacing the extension .pdb with .prepi and .frcmod
The antechamber and parmchk program should exist in the system path or the AMBERHOME environment variable should be set correctly.
build_template.py¶
Program to build a ProtoMS template file
usage: build_template.py [-h] [-p PREPI] [-o OUT] [-z ZMAT] [-f FRCMOD]
[-n NAME] [-t TRANSLATE] [-r ROTATE] [--alldihs]
[--gaff GAFF]
Named Arguments¶
| -p, --prepi | the name of the leap prepi-file |
| -o, --out | the name of the template file Default: “lig.tem” |
| -z, --zmat | the name of the zmatrix-file, if it exists |
| -f, --frcmod | the name of the frcmod-file, if it exists |
| -n, --name | the name of the solute Default: “UNK” |
| -t, --translate | |
maxmium size for translation moves in Angstroms Default: 0.1 | |
| -r, --rotate | maxmium size for rotation moves in degrees Default: 1.0 |
| --alldihs | sample improper dihedrals Default: False |
| --gaff | gaff version to use, gaff14 or gafff16 Default: “gaff16” |
Examples:
build_template.py -p benzene.prepi
build_template.py -p benzene.prepi -f benzene.frcmod
build_template.py -p benzene.prepi -f benzene.frcmod -o benzene.template -n BNZ
build_template.py -p benzene.prepi -f benzene.frcmod -t 1.0 -r 10
Description:
This tool builds a ProtoMS template file for a solute given an Amber prepi file.
If the solute needs parameters not in the specified GAFF release, they should be supplied with the frcmodfile.
The tool will automatically make an appropriate z-matrix for Monte Carlo sampling. This works in most situations. However, if something is not working properly with the generated z-matrix, one can be supplied in the zmatfile
The default translational and rotational displacements are based on experience and should be appropriate in most situations.
calc_clusters.py¶
usage: calc_clusters.py [-h] [-i INPUT] [-m MOLECULE] [-a ATOM] [-s SKIP]
[-c CUTOFF] [-l LINKAGE] [-o OUTPUT]
Named Arguments¶
| -i, --input | PDB file containing input frames. Default=’all.pdb’ Default: “all.pdb” |
| -m, --molecule | Residue name of water molecules. Default=’WA1’ Default: “WA1” |
| -a, --atom | Name of atom to take as molecule coordinates. Default=’O00’ Default: “O00” |
| -s, --skip | Number of frames to skip. Default=0 Default: 0 |
| -c, --cutoff | Distance cutoff for clustering. Default=2.4 Angs Default: 2.4 |
| -l, --linkage | Linkage method for hierarchical clustering. Default=’average’ Default: “average” |
| -o, --output | Filename for the PDB output. Default=’clusters.pdb’ Default: “clusters.pdb” |
Examples:
calc_clusters.py -i all.pdb
calc_clusters.py -i all.pdb all2.pdb
calc_clusters.py -i all.pdb -o all_clusters.pdb
calc_clusters.py -i all.pdb -t complete
Description:
This tool cluster molecules from a simulation
It will extract the coordinates of all atoms with name equal to atom in residues with name equal to molecule in all input files and cluster them using the selected algorithm. If no atom is specified, the entire molecule will be clustered. By default this atom and residue name is set to match GCMC / JAWS output with the standard water template.
calc_density.py¶
Program to discretize atoms on a 3D grid
usage: calc_density.py [-h] [-f FILES [FILES ...]] [-o OUT] [-r RESIDUE]
[-a ATOM] [-p PADDING] [-s SPACING] [-e EXTENT]
[-n NORM] [-t {sphere,gaussian}] [--skip SKIP]
[--max MAX]
Named Arguments¶
| -f, --files | the input PDB-files |
| -o, --out | the name of the output grid-file in DX-format, default=’grid.dx’ Default: “grid.dx” |
| -r, --residue | the name of the residue to extract, default=’wa1’ Default: “wa1” |
| -a, --atom | the name of the atom to extract, default=’o00’ Default: “o00” |
| -p, --padding | the amount to increase the minimum box in each direction, default=2 A Default: 2.0 |
| -s, --spacing | the grid resolution, default=0.5 A Default: 0.5 |
| -e, --extent | the size of the smoothing, i.e. the extent of an atom, default=1A Default: 1.0 |
| -n, --norm | number used to normalize the grid, if not specified the number of input files is used |
| -t, --type | Possible choices: sphere, gaussian the type of coordinate smoothing, should be either ‘sphere’, ‘gaussian’ Default: “sphere” |
| --skip | the number of blocks to skip to calculate the density. default is 0. Skip must be greater or equal to 0 Default: 0 |
| --max | the upper block to use. default is 99999 which should make sure you will use all the available blocks. max must be greater or equal to 0 Default: 99999 |
Examples:
calc_density.py -i all.pdb
calc_density.py -i all.pdb all2.pdb
calc_density.py -i all.pdb -o gcmc_density.dx
calc_density.py -i all.pdb -r t4p -n o00
calc_density.py -i all.pdb -p 1.0 -s 1.0
calc_density.py -i all.pdb -e 0.5 -t gaussian
calc_density.py -i all.pdb -n 100
Description:
This tool discretises atoms on a grid, thereby representing a simulation output as a density.
It will extract the coordinates of all atoms with name equal to atom in residues with name equal to residue in all input files and discretise them on a grid. By default this atom and residue name is set to match GCMC / JAWS output with the standard water template.
The produced density can be visualized with most programs, e.g.
vmd -m all.pdb grid.dx
calc_dg.py¶
Calculate free energy differences using a range of estimators
usage: calc_dg.py [-h] -d DIRECTORIES [DIRECTORIES ...] [-l LOWER_BOUND]
[-u UPPER_BOUND] [-t TEMPERATURE] [--pickle PICKLE]
[--save-figures [SAVE_FIGURES]] [--no-show] [-n NAME]
[--subdir SUBDIR] [--pmf]
[--test-equilibration TEST_EQUILIBRATION]
[--test-convergence TEST_CONVERGENCE]
[--estimators {ti,mbar,bar,gcap} [{ti,mbar,bar,gcap} ...]]
[-v VOLUME]
Named Arguments¶
| -d, --directories | |
| Location of folders containing ProtoMS output subdirectories. Multiple directories can be supplied to this flag and indicate repeats of the same calculation. This flag may be given multiple times and each instances is treated as an individual leg making up a single free energy difference e.g. vdw and ele contributions of a single topology calculation. | |
| -l, --lower-bound | |
Value between 0 and 1 that determines the proportion to omit from the beginning of the simulation data series. Default: 0.0 | |
| -u, --upper-bound | |
Value between 0 and 1 that determines the proportion to omit from the end of the simulation data series. Default: 1.0 | |
| -t, --temperature | |
Temperature at which the simulation was run. Default=298.15K Default: 298.15 | |
| --pickle | Name of file in which to store results as a pickle. |
| --save-figures | Save figures produced by script. Takes optional argument that adds a prefix to figure names. |
| --no-show | Do not display any figures on screen. Does not interfere with –save-figures. Default: False |
| -n, --name | Name of ProtoMS output file containing free energy data. Note that this option will not change the output file used by the gcap estimator from results_inst. Default: “results” |
| --subdir | Optional sub-directory for each lambda value to search within for simulation output. This is useful in, for instance, processing only the results of a GCAP calculation at a particular B value. Default: “” |
| --pmf | Make graph of potential of mean force Default: False |
| --test-equilibration | |
| Perform free energy calculations 10 times using varying proportions of the total data set provided. Data used will range from 100% of the dataset down to the proportion provided to this argument | |
| --test-convergence | |
| Perform free energy calculations 10 times using varying proportions of the total data set provided. Data used will range from 100% of the dataset up to the proportion provided to this argument | |
| --estimators | Possible choices: ti, mbar, bar, gcap Choose free energy estimator to use. By default TI, BAR and MBAR are used. Note that the GCAP estimator assumes a different file structure and ignores the –subdir flag. Default: [‘ti’, ‘mbar’, ‘bar’] |
| -v, --volume | Volume of GCMC region |
Examples:
calc_dg.py -d out_free/
calc_dg.py -d out_free1/ out_free2/ out_free3/ -l 0.1
calc_dg.py -d out_free1/ out_free2/ out_free3/ -u 0.9
calc_dg.py -d out_free1/ out_free2/ out_free3/ -e ti bar
calc_dg.py -d out_free1/ out_free2/ out_free3/ -e gcap
calc_dg.py -d out_free1/ out_free2/ out_free3/ --subdir b_-9.700 -e ti bar
Description:
This tool calculates free energies using the method of thermodynamic integration (TI), Bennett’s Acceptance Ratio (BAR), multi state BAR (MBAR) and grand canonical alchemical perturbation (GCAP).
The program expects that in the directory, directory2 etc. there exists an output folder for each  -value, eg.
-value, eg. lam-0.000 and lam-1.000 (unless the --subdir argument is used.)
calc_dg_cycle.py¶
High level script that attempts to use data from multiple calculations to provide free energies of solvation and binding. Also calculates cycle closures for all data. Assumes standard ProtoMS naming conventions for data output directories. Data should be organised such that each transformation between two ligands should have a single master directory containing output directories for each simulation state (e.g. master/out1_free). Masterdirectories should be passed to the -d flag. Reported free energies are averages over all repeats found. Reported errors are single standard errors calculated from repeats.
usage: calc_dg_cycle.py [-h] [-l LOWER_BOUND] [-u UPPER_BOUND]
[-t TEMPERATURE] [--pickle PICKLE]
[--save-figures [SAVE_FIGURES]] [--no-show] [-n NAME]
[--subdir SUBDIR] -d DIRECTORIES [DIRECTORIES ...] -s
{+,-} [{+,-} ...]
[--estimators {ti,mbar,bar,gcap} [{ti,mbar,bar,gcap} ...]]
[-v VOLUME]
(--dualtopology | --singletopology {comb,sep})
Named Arguments¶
| -l, --lower-bound | |
Value between 0 and 1 that determines the proportion to omit from the beginning of the simulation data series. Default: 0.0 | |
| -u, --upper-bound | |
Value between 0 and 1 that determines the proportion to omit from the end of the simulation data series. Default: 1.0 | |
| -t, --temperature | |
Temperature at which the simulation was run. Default=298.15K Default: 298.15 | |
| --pickle | Name of file in which to store results as a pickle. |
| --save-figures | Save figures produced by script. Takes optional argument that adds a prefix to figure names. |
| --no-show | Do not display any figures on screen. Does not interfere with –save-figures. Default: False |
| -n, --name | Name of ProtoMS output file containing free energy data. Note that this option will not change the output file used by the gcap estimator from results_inst. Default: “results” |
| --subdir | Optional sub-directory for each lambda value to search within for simulation output. This is useful in, for instance, processing only the results of a GCAP calculation at a particular B value. Default: “” |
| -d, --directories | |
| Location of folders containing ProtoMS output directories. | |
| -s, --signs | Possible choices: +, - List of ‘+’ or ‘-‘ characters, one for each directory provided to the -d flag. Indicates the sign that should be used for each free energy difference when calculating cycle closures. |
| --estimators | Possible choices: ti, mbar, bar, gcap Choose estimators Default: [‘ti’, ‘mbar’, ‘bar’] |
| -v, --volume | Volume of GCMC region |
| --dualtopology | Indicates data is for a dual topology calculation. Default: False |
| --singletopology | |
Possible choices: comb, sep Indicates data is for a single topology calculation. Option comb indicates a single step calculation. Option sep indicates separate steps for van der Waals and electrostatics components. | |
Examples:
calc_dg_cycle.py -d a_b/out_free -s + b_c/out_free -s + a_c/out_free -s - --dualtopology
calc_dg_cycle.py -d a_b/out_free -s + b_c/out_free -s + a_c/out_free -s - --singletopology comb
Description:
Calculates thermodynamic cycle closure for a set of simulations. This can be performed either for dual topology results, or single topology results. With single topology simulations, the electrostatic and van der Waals results can either be considered separately --singletopology sep or together, --singletopology comb.
calc_gcap_surface.py¶
Calculate free energy differences using a range of estimators
usage: calc_gcap_surface.py [-h] -d DIRECTORIES [DIRECTORIES ...]
[-l LOWER_BOUND] [-u UPPER_BOUND] [-t TEMPERATURE]
[--pickle PICKLE] [--save-figures [SAVE_FIGURES]]
[--no-show] [-n NAME] [--subdir SUBDIR]
[-v VOLUME]
[--estimators {ti,mbar,bar} [{ti,mbar,bar} ...]]
Named Arguments¶
| -d, --directories | |
| Location of folders containing ProtoMS output subdirectories. Multiple directories can be supplied to this flag and indicate repeats of the same calculation. This flag may be given multiple times and each instances is treated as an individual leg making up a single free energy difference e.g. vdw and ele contributions of a single topology calculation. | |
| -l, --lower-bound | |
Value between 0 and 1 that determines the proportion to omit from the beginning of the simulation data series. Default: 0.0 | |
| -u, --upper-bound | |
Value between 0 and 1 that determines the proportion to omit from the end of the simulation data series. Default: 1.0 | |
| -t, --temperature | |
Temperature at which the simulation was run. Default=298.15K Default: 298.15 | |
| --pickle | Name of file in which to store results as a pickle. |
| --save-figures | Save figures produced by script. Takes optional argument that adds a prefix to figure names. |
| --no-show | Do not display any figures on screen. Does not interfere with –save-figures. Default: False |
| -n, --name | Name of ProtoMS output file containing free energy data. Note that this option will not change the output file used by the gcap estimator from results_inst. Default: “results” |
| --subdir | Optional sub-directory for each lambda value to search within for simulation output. This is useful in, for instance, processing only the results of a GCAP calculation at a particular B value. Default: “” |
| -v, --volume | Volume of GCMC region |
| --estimators | Possible choices: ti, mbar, bar Choose free energy estimator to use. By default TI, BAR and MBAR are used. Note that the GCAP estimator assumes a different file structure and ignores the –subdir flag. Default: [‘ti’, ‘mbar’, ‘bar’] |
Examples:
calc_gcap_surface.py -d out_gcap -v 300.
calc_gcap_surface.py -d out_gcap --save-figures -v 300.
calc_gcap_surface --subdir b_-9.700 --estimators mbar -v 300.
Description:
Calculates the free energy from a surface-GCAP simulation. The volume of the GCMC region must be given using the -v flag. To calculate the free energy at a single B value, use the --subdir flag with calc_dg.py, and the energy can be calculated with any one dimensional free energy method.
calc_gci.py¶
Calculate water binding free energies using Grand Canonical Integration.
usage: calc_gci.py [-h] [-l LOWER_BOUND] [-u UPPER_BOUND] [-t TEMPERATURE]
[--pickle PICKLE] [--save-figures [SAVE_FIGURES]]
[--no-show] [--name NAME] -d DIRECTORIES [DIRECTORIES ...]
-v VOLUME [-n NSTEPS] [--nmin NMIN] [--nmax NMAX]
[--nfits NFITS] [--pin_min PIN_MIN]
Named Arguments¶
| -l, --lower-bound | |
Value between 0 and 1 that determines the proportion to omit from the beginning of the simulation data series. Default: 0.0 | |
| -u, --upper-bound | |
Value between 0 and 1 that determines the proportion to omit from the end of the simulation data series. Default: 1.0 | |
| -t, --temperature | |
Temperature at which the simulation was run. Default=298.15K Default: 298.15 | |
| --pickle | Name of file in which to store results as a pickle. |
| --save-figures | Save figures produced by script. Takes optional argument that adds a prefix to figure names. |
| --no-show | Do not display any figures on screen. Does not interfere with –save-figures. Default: False |
| --name | Name of ProtoMS output file containing free energy data. Note that this option will not change the output file used by the gcap estimator from results_inst. Default: “results” |
| -d, --directories | |
| Location of folders containing ProtoMS output subdirectories. Multiple directories can be supplied to this flag and indicate repeats of the same calculation. | |
| -v, --volume | Volume of the calculations GCMC region. |
| -n, --nsteps | Override automatic guessing of the number of steps to fit for titration curve fitting. |
| --nmin | Override automatic guessing of the minimum number of waters for tittration curve fitting. |
| --nmax | Override automatic guessing of maximum number of waters for titration curve fitting. |
| --nfits | The number of independent fitting attempts for the neural network occupancy model. Increasing the number of fits may help improve results for noisy data. Default: 10 |
| --pin_min | The minimum value when fitting the neural network occupancy model. Setting this may help improve models which are poorly fit at low values |
Examples:
calc_gci.py -d out_gcmc/ -v 130.
calc_gci.py -d out_gcmc/ -v 130. -l 0.2
calc_gci.py -d out_gcmc/ -v 130. --save-figures
calc_gci.py -d out_gcmc/ -v 130. --pin_min
Description:
Collection of tools to analyse and visualise GCMC titration data of water using grand canonical integration (GCI). Used to plot average number of waters for a given Adams value, i.e. GCMC titration data, calculate transfer free energies from ideal gas, calculate absolute and relative binding free energies of water, calculate and/or estimate optimal number of bound waters. As described in Ross et al., J. Am. Chem. Soc., 2015, 137 (47), pp 14930-14943.
Error estimates of free energies and optimal number of waters are based on automatic repeated fitting of the ANN from different random initial parameters. This can be increased with --nfits.
calc_replicapath.py¶
Program to analyze and plot a replica paths
usage: calc_replicapath.py [-h] [-f FILES [FILES ...]] [-p PLOT [PLOT ...]]
[-k {lambda,temperature,rest,global,B}] [-o OUT]
Named Arguments¶
| -f, --files | the name of the files to analyse |
| -p, --plot | the replica values to plot |
| -k, --kind | Possible choices: lambda, temperature, rest, global, B the kind of replica to analyze Default: “lambda” |
| -o, --out | the prefix of the output figure. Default is replica_path. Default: “replica_path.png” |
Examples:
calc_replicapath.py -f out_free/lam-0.*/results -p 0.000 1.000
calc_replicapath.py -f out_free/lam-0.*/results -p 0.000 0.500 1.000 -o replica_paths.png
calc_replicapath.py -f out_free/t-*/lam-0.000/results -p 25.0 35.0 45.0 -k temperature
Description:
This tools plots the path of different replicas in a replica exchange simulation as a function of simulation time.
If the kind of replicas is from  replica exchange the
replica exchange the replica1 and replica2 etc should be individual  -values to plot.
-values to plot.
If the kind of replicas is from REST or temperature replica exchange the replica1 and replica2 etc should be individual temperatures to plot.
calc_rmsd.py¶
Program to calculate RMSD of ligand centre
usage: calc_rmsd.py [-h] [-i INITIAL] [-f FILES [FILES ...]] [-l LIGAND]
[-a ATOM] [-t TEMPERATURE]
Named Arguments¶
| -i, --initial | the initial PDB-file of the ligand |
| -f, --files | the input PDB-files |
| -l, --ligand | the name of the ligand to extract |
| -a, --atom | the name of the atom to analyze |
| -t, --temperature | |
the temperature in the simulation Default: 298.0 | |
Examples:
calc_rmsd.py -i benzene.pdb -f out_bnd/all.pdb -r bnz
calc_rmsd.py -i benzene.pdb -f out_bnd/all.pdb -r bnz -a c4
Description:
This tool calculate the RMSD of a ligand in a simulation.
If the atom name is given, the tool will calculate the RMSD of that atom with respect to its position in pdbfile. Otherwise, the program will calculate the RMSD of the geometric centre with respect to pdbfile.
A force constant to keep the ligand restrained for free energy calculations is estimated from the RMSD using the equipartition theorem.
calc_series.py¶
Program to analyze and plot a time series
usage: calc_series.py [-h] [-f FILE [FILE ...]] [-o OUT]
[-s SERIES [SERIES ...]]
[-p {sep,sub,single,single_first0,single_last0}]
[--nperm NPERM] [--threshold THRESHOLD] [--average]
[--moving MOVING]
Named Arguments¶
| -f, --file | the name of the file to analyse. Default is results. Default: [‘results’] |
| -o, --out | the prefix of the output figure. Default is series. Default: “series” |
| -s, --series | the series to analyze |
| -p, --plot | Possible choices: sep, sub, single, single_first0, single_last0 the type of plot to generate for several series |
| --nperm | if larger than zero, perform a permutation test to determine equilibration, default=0 Default: 0 |
| --threshold | the significant level of the equilibration test, default=0.05 Default: 0.05 |
| --average | turns on use of running averaging of series Default: False |
| --moving | turns on use of moving averaging of series, default=None |
The tool will estimate the number of independent samples for a given observable in the production part using the method of statistical inefficiency. The equilibration time will also be estimated from a method that maximizes the number uncorrelated samples as suggested on alchemistry.org.
Apart from the raw series, the tool can also plot the running average if the --average flag is set or the moving average if the --moving flag is used.
Typically only a single ProtoMS results file will be analysed and plotted. However, for the series grad and agrad (the gradient and analytical gradient, respectively), multiple results file can be given. In this case, the gradients for each results file is used to estimate the free energy using thermodynamic integration.
calc_ti_decomposed.py¶
Calculate individual contributions of different terms to the total free energy difference. Although terms are guaranteed to be additive with TI, the decomposition is not strictly well defined. That said, it can be illustrative to consider the dominant contributions of a calculation.
usage: calc_ti_decomposed.py [-h] -d DIRECTORIES [DIRECTORIES ...]
[-l LOWER_BOUND] [-u UPPER_BOUND]
[-t TEMPERATURE] [--pickle PICKLE]
[--save-figures [SAVE_FIGURES]] [--no-show]
[-n NAME] [--subdir SUBDIR]
[-b BOUND [BOUND ...]] [-g GAS [GAS ...]]
[--dualtopology] [--pmf] [--full]
Named Arguments¶
| -d, --directories | |
| Location of folders containing ProtoMS output subdirectories. Multiple directories can be supplied to this flag and indicate repeats of the same calculation. This flag may be given multiple times and each instances is treated as an individual leg making up a single free energy difference e.g. vdw and ele contributions of a single topology calculation. | |
| -l, --lower-bound | |
Value between 0 and 1 that determines the proportion to omit from the beginning of the simulation data series. Default: 0.0 | |
| -u, --upper-bound | |
Value between 0 and 1 that determines the proportion to omit from the end of the simulation data series. Default: 1.0 | |
| -t, --temperature | |
Temperature at which the simulation was run. Default=298.15K Default: 298.15 | |
| --pickle | Name of file in which to store results as a pickle. |
| --save-figures | Save figures produced by script. Takes optional argument that adds a prefix to figure names. |
| --no-show | Do not display any figures on screen. Does not interfere with –save-figures. Default: False |
| -n, --name | Name of ProtoMS output file containing free energy data. Note that this option will not change the output file used by the gcap estimator from results_inst. Default: “results” |
| --subdir | Optional sub-directory for each lambda value to search within for simulation output. This is useful in, for instance, processing only the results of a GCAP calculation at a particular B value. Default: “” |
| -b, --bound | Output directory(s) of additional bound phase calculation(s). Using this flag causes data loaded via -d to be considered as solvent phase data. All data is then combined to provide a decomposition of the binding free energy. Behaves identically to -d in treatment of repeats and calculation legs. |
| -g, --gas | As -b except data loaded via this flag is treated as gas phase data to provide to provide a decomposed solvation free energy. |
| --dualtopology | Indicates provided data is from a dual topology calculation. Attempts to consolidate terms, for clarity, from ligands that can have opposite signs and large magnitudes. Please note that standard errors calculated with this approach are no longer rigorous and can be spuriously large. Default: False |
| --pmf | Plot the Potential of Mean Force for all terms. Default: False |
| --full | Prevents printing out of zero contribution energies. Default: True |
Examples:
calc_ti.py -d out_free/
calc_ti.py -d out_free/ -l 0.1 -u 0.9
calc_ti.py -b out_bnd/ -d out_free --dualtopology
calc_ti.py -d out_free -g out_gas
Description:
This tool calculates free energies of individual energetic components using the method of thermodynamic integration (TI).
The program expects that in the directory there exist an output folder for each  -value, eg.
-value, eg. lam-0.000 and lam-1.000
Block estimates can be constructed by combining -l and -u. For instance, these commands calculates the free energy while incrementally increasing the equilibration
for X in `seq 0.0 0.1 1.0`
do
calc_ti_decomposed.py -d out_free -l $x
done
clear_gcmcbox.py¶
Program to remove water molecules from a GCMC/JAWS-1 box
usage: clear_gcmcbox.py [-h] [-b BOX] [-s SOLVATION] [-o OUT]
Named Arguments¶
| -b, --box | the name of the PDB-file containing the box. |
| -s, --solvation | |
| the name of the PDB-file containing the solvation waters | |
| -o, --out | the name of the output PDB-file Default: “cleared_box.pdb” |
Examples:
clear_gcmcbox.py -b gcmc_box.pdb -s water.pdb
clear_gcmcbox.py -b gcmc_box.pdb -s water.pdb -o water_cleared.pdb
Description:
This tool clears a GCMC or JAWS-1 simulation box from any bulk water placed there by the solvation method.
In a GCMC and JAWS-1 simulation the bulk water is prevented to enter or exit a GCMC or JAWS-1 simulation box. Therefore, bulk water that are within this box needs to be removed prior to the GCMC or JAWS-1 simulation.
The boxfile is typically created by make_gcmcbox.py and the waterfile is typically created by solvate.py and can be either a droplet or a box.
convertatomnames.py¶
Program convert atom names in a protein pdb-file to ProtoMS style
usage: convertatomnames.py [-h] [-p PROTEIN] [-o OUT] [-s STYLE]
[-c CONVERSIONFILE]
Named Arguments¶
| -p, --protein | the protein PDB-file |
| -o, --out | the output PDB-file Default: “protein_pms.pdb” |
| -s, --style | the style of the input PDB-file Default: “amber” |
| -c, --conversionfile | |
the name of the file with conversion rules Default: “atomnamesmap.dat” | |
Examples:
convertatomnames.py -p protein.pdb
convertatomnames.py -p protein.pdb -c $PROTOMSHOME/data/atomnamesmap.dat
convertatomnames.py -p protein.pdb -s charmm
Description:
This tool converts residue and atom names to ProtoMS convention.
This script modfies in particular names of hydrogen atoms, but also some residue names, e.g. histidines.
A file containing conversion instructions for amber and charmm is available in the $PROTOMSHOME/data folder.
convertwater.py¶
Program to convert water molecules - with or without hydrogens - in a pdb file to simulation models, such as tip4p. Currently ignores original hydrogen positions.
usage: convertwater.py [-h] [-p PDB] [-o OUT] [-m MODEL] [-i] [-n RESNAME]
[--setupseed SETUPSEED]
Named Arguments¶
| -p, --pdb | the PDF-file containing the waters to be transformed |
| -o, --out | the output PDB-file Default: “convertedwater.pdb” |
| -m, --model | the water model,default=tip4p Default: “tip4p” |
| -i, --ignoreh | whether to ignore hydrogens in input water. If no hydrogens are present, waters are randomly orientated. default=No Default: False |
| -n, --resname | the residue name that will be applied to the water molecules. When it is not specified, it is chosen based on the water model |
| --setupseed | optional random number seed for generation of water coordinates |
Examples:
convertwater.py -p protein.pdb
convertwater.py -p protein.pdb -m tip3p
convertwater.py -p protein.pdb --ignoreh
Description:
This tool converts water molecules to a specific model.
Currently the script recognizes TIP3P and TIP4P water models. The valid values for style is therefore t4p, tip4p, tp4, t3p, tip3p, tp3
If the --ignoreh flag is given, the script will discard the hydrogen atoms found in pdbfile and add them at a random orientation.
distribute_waters.py¶
Randomly distribute n molecules within box dimensions
usage: distribute_waters.py [-h] [-b BOX BOX BOX BOX BOX BOX] [-m MOLECULES]
[-o OUTFILE] [--model MODEL] [--resname RESNAME]
[--number NUMBER] [--setupseed SETUPSEED]
Named Arguments¶
| -b, --box | Dimensions of the box. Six arguments expected: origin (x,y,z) & length (x,y,z) |
| -m, --molecules | |
| Molecules to distribute in the box. Either the number of waters or a pdb file containing all of them | |
| -o, --outfile | Name of the pdb file to write the molecules to. Default=’ghostmolecules.pdb’ Default: “ghostmolecules.pdb” |
| --model | Water model. Used when only the amount of waters is specified. Options: ‘t4p’,’t3p’. Default=’t4p’ Default: “t4p” |
| --resname | Residue name of the molecules writen to output. Default=’WAT’ Default: “WAT” |
| --number | Required number of molecules when it differs from the number of residues in the file. |
| --setupseed | Optional random number seed for generation of water coordinates |
Examples:
distribute_waters.py -b 53.4 56.28 13.23 10 10 10 -m 12
distribute_waters.py -b 53.4 56.28 13.23 10 10 10 -m 12 --model t3p --resname T3P
distribute_waters.py -b 53.4 56.28 13.23 10 10 10 -m myonewater.pdb --number 12 -o mywatersinbox.pdb
Description:
This tool can place water molecules at random within a GCMC or JAWS-1 simulation box.
It can place molecules in random positions and orientations with their geometry center restricted to the given dimensions of a box.
divide_pdb.py¶
Split your multi pdb file into individual files
usage: divide_pdb.py [-h] [-i INPUT] [-o OUTPUT] [-p PATH]
Named Arguments¶
| -i, --input | The name of your multi pdb file. Default = all.pdb Default: “all.pdb” |
| -o, --output | The basename of your individual pdb files. Default = snapshot_ Default: “snapshot_” |
| -p, --path | Where the input should be found and the output printed. Default = ./ Default: “./” |
Examples:
- ::
- divide_pdb.py divide_pdb.py -i mypmsout.pdb -o individual -p outfolder/
Description:
This tool splits up a PDB file with multiple models (the keyword END defines the end of a model) into several PDB files.
generate_input.py¶
Program to create a ProtoMS command file
usage: generate_input.py [-h]
[-s {sampling,equilibration,dualtopology,singletopology,gcap_single,gcap_dual,gcmc,jaws1,jaws2}]
[--dovacuum] [-p PROTEIN] [-l LIGANDS [LIGANDS ...]]
[-t TEMPLATES [TEMPLATES ...]] [-pw PROTWATER]
[-lw LIGWATER] [-o OUT] [--outfolder OUTFOLDER]
[--gaff GAFF] [--lambdas LAMBDAS [LAMBDAS ...]]
[--adams ADAMS [ADAMS ...]]
[--adamsrange ADAMSRANGE [ADAMSRANGE ...]]
[--jawsbias JAWSBIAS [JAWSBIAS ...]]
[--gcmcwater GCMCWATER] [--gcmcbox GCMCBOX]
[--watmodel {tip3p,tip4p}] [--nequil NEQUIL]
[--nprod NPROD] [--dumpfreq DUMPFREQ] [--absolute]
[--ranseed RANSEED]
[--softcore {auto,all,none,manual}]
[--spec-softcore SPEC_SOFTCORE]
Named Arguments¶
| -s, --simulation | |
Possible choices: sampling, equilibration, dualtopology, singletopology, gcap_single, gcap_dual, gcmc, jaws1, jaws2 the kind of simulation to setup Default: “equilibration” | |
| --dovacuum | turn on vacuum simulation for simulation types equilibration and sampling Default: False |
| -p, --protein | the name of the protein file |
| -l, --ligands | the name of the ligand pdb files |
| -t, --templates | |
| the name of ProtoMS template files | |
| -pw, --protwater | |
| the name of the solvent for protein | |
| -lw, --ligwater | |
| the name of the solvent for ligand | |
| -o, --out | the prefix of the name of the command file Default: “run” |
| --outfolder | the ProtoMS output folder Default: “out” |
| --gaff | the version of GAFF to use for ligand Default: “gaff16” |
| --lambdas | the lambda values or the number of lambdas Default: [16] |
| --adams | the Adam/B values for the GCMC Default: 0 |
| --adamsrange | the upper and lower Adam/B values for the GCMC and, optionally, the number of values desired (default value every 1.0), e.g. -1 -16 gives all integers between and including -1 and -16 |
| --jawsbias | the bias for the JAWS-2 Default: 0 |
| --gcmcwater | a pdb file with a box of water to do GCMC on |
| --gcmcbox | a pdb file with box dimensions for the GCMC box |
| --watmodel | Possible choices: tip3p, tip4p the name of the water model. Default = tip4p Default: “tip4p” |
| --nequil | the number of equilibration steps Default: 5000000.0 |
| --nprod | the number of production steps Default: 40000000.0 |
| --dumpfreq | the output dump frequency Default: 100000.0 |
| --absolute | whether an absolute free energy calculation is to be run. Default=False Default: False |
| --ranseed | the value of the random seed you wish to simulate with. If None, then a seed is randomly generated. Default=None |
| --softcore | Possible choices: auto, all, none, manual determine which atoms to apply softcore potentials to. If ‘all’ softcores are applied to all atoms of both solutes. If ‘none’ softcores are not applied to any atoms. If ‘auto’, softcores are applied to atoms based on matching coordinates between ligand structures. The selected softcore atoms can be amended using the –spec-softcore flag. If ‘manual’ only those atoms specified by the –spec-softcore flag are softcore. Default: “all” |
| --spec-softcore | |
| Specify atoms to add or remove from softcore selections. Can be up to two, space separated, strings of the form “N:AT1,AT2,-AT3”. N should be either “1” or “2” indicating the corresponding ligand. The comma separated list of atom names are added to the softcore selection. A preceding dash for an atom name specifies it should be removed from the softcore selection. The special value “auto” indictates that automatic softcore assignments should be accepted without amendment. | |
Examples:
generate_input.py -s dualtopology -l lig1.pdb lig2.pdb -p protein.pdb -t li1-li2.tem -pw droplet.pdb -lw lig1_wat.pdb --lambas 8
generate_input.py -s dualtopology -l lig1.pdb dummy.pdb -t li1-dummy.tem -lw lig1_wat.pdb --absolute
generate_input.py -s gcmc -p protein.pdb -pw droplet.pdb --adams -4 -2 0 2 4 6 --gcmcwater gcmc_water.pdb --gcmcbox gcmc_box.pdb
generate_input.py -s sampling -l lig1.pdb -t lig1.tem --dovacuum
Description:
This tool generates input files with commands for ProtoMS.
The settings generate are made according to experience and should work in most situations.
The tool will create at most two ProtoMS command files, one for the protein simulation and one for the ligand simulation. These can be used to run ProtoMS, e.g.
$PROTOMS/protoms3 run_free.cmd
make_dummy.py¶
Program make a dummy corresponding to a molecule
usage: make_dummy.py [-h] [-f FILE] [-o OUT]
Named Arguments¶
| -f, --file | the name of a PDB file |
| -o, --out | the name of the dummy PDB file Default: “dummy.pdb” |
Examples:
make_dummy.py -f benzene.pdb
make_dummy.py -f benzene.pdb -o benzene_dummy.pdb
Description:
This tool makes a matching dummy particle for a solute.
The dummy particle will be placed at the centre of the solute.
make_gcmcbox.py¶
Program to make a PDB-file with box coordinates covering a solute molecules
usage: make_gcmcbox.py [-h] [-s SOLUTE] [-p PADDING] [-o OUT]
[-b BOX [BOX ...]]
Named Arguments¶
| -s, --solute | the name of the PDB-file containing the solute. |
| -p, --padding | the padding in A,default=2 Default: 2.0 |
| -o, --out | the name of the box PDB-file Default: “gcmc_box.pdb” |
| -b, --box | Either the centre of the box (x,y,z), or the centre of box AND length (x,y,z,x,y,z). If the centre is specified and the length isn’t, twice the ‘padding’ will be the lengths of a cubic box. |
Examples:
make_gcmcbox.py -s benzene.pdb
make_gcmcbox.py -s benzene.pdb -p 0.0
make_gcmcbox.py -s benzene.pdb -o benzene_gcmc_box.pdb
Description:
This tool makes a GCMC or JAWS-1 simulation box to fit on top of a solute.
The box will be created so that it has the extreme dimensions of the solute and then padding will be added in each dimension
The box can be visualised with most common programs, e.g.
vmd -m benzene.pdb benzene_gcmc_box.pdb
this is a good way to see that the box is of appropriate dimensions.
When an appropriate box has been made, it can be used by solvate.py to fill it with water.
make_single.py¶
Program to setup template files for single-toplogy perturbations semi-automatically
usage: make_single.py [-h] [-t0 TEM0] [-t1 TEM1] [-p0 PDB0] [-p1 PDB1]
[-m MAP] [-o OUT] [--gaff GAFF]
Named Arguments¶
| -t0, --tem0 | Template file for V0 |
| -t1, --tem1 | Template file for V1 |
| -p0, --pdb0 | PDB-file for V0 |
| -p1, --pdb1 | PDB-file for V1 |
| -m, --map | the correspondance map from V0 to V1 |
| -o, --out | prefix of the output file Default: “single” |
| --gaff | the version of GAFF to use for ligand Default: “gaff16” |
Examples:
make_single.py -t0 benzene.tem -t1 toluene.tem -p0 benzene.pdb -p1 toluene.pdb
make_single.py -t0 benzene.tem -t1 toluene.tem -p0 benzene.pdb -p1 toluene.pdb -m bnz2tol.dat
make_single.py -t0 benzene.tem -t1 toluene.tem -p0 benzene.pdb -p1 toluene.pdb -o bnz-tol
Description:
This tool makes ProtoMS template files for single topology free energy simulations.
The program will automatically try to match atoms in template0 with atoms in template1. It will do this by looking for atoms with the same atom type that are on top of each other in pdbfile0 and pdbfile1. A cut-off of 0.02 A2 will be used for this. All atoms that cannot be identified in this way are written to the screen and the user has to enter the corresponding atoms. If no corresponding atom exists, i.e., the atom should be perturbed to a dummy, the user may enter blank.
The user may also write the corresponding atoms to a file and provide it as map above. In this file there should be one atom pair on each line, separated by white-space. A dummy atom should be denoted as DUM. If map is not given, the program will write the created correspondence map to a file based on the outfile string.
Currently, dummy atoms are not supported in the solute at 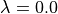 . Therefore, this solute needs to be the larger one.
. Therefore, this solute needs to be the larger one.
The tool will write two ProtoMS template files, one for the electrostatic perturbation, one for the van der Waals perturbation and one for the combined perturbation. These template files will end in _ele.tem, _vdw.tem, _comb.tem respectively.
A summary of the charges and van der Waals parameters in the four states will be printed to the screen. This information should be checked carefully.
merge_templates.py¶
Program merge a series of ProtoMS template files
usage: make_templates.py [-h] [-f FILES [FILES ...]] [-o OUT]
Named Arguments¶
| -f, --files | the name of the template files |
| -o, --out | the name of the merged template file |
Examples:
merge_templates.py -f benzene.tem dummy.tem -o bnz-dummy.tem
Description:
This tool combines several ProtoMS template files into a single template file.
The force field parameters in file2 will be re-numbered so that they do not conflict with file1. This is important when you want to load both parameters into ProtoMS at the same time.
plot_theta.py¶
Program to plot the theta distribution of a given molecule, result from a JAWS simulation
usage: plot_theta.py [-h] [-r RESULTS] [-s RESTART] [-m MOLECULE]
[-p PLOTNAME] [--skip SKIP]
Named Arguments¶
| -r, --results | the name of the results file. Deafult=’results’ Default: “results” |
| -s, --restart | the replica values to plot. Default=’restart’ Default: “restart” |
| -m, --molecule | the residue name of the JAWS molecule. Default=’WAT’ Default: “WAT” |
| -p, --plotname | the start of the filename for the plots generated. Default=’theta_dist’ Default: “theta_dist” |
| --skip | the number of results snapshots to skip, Default = 0 Default: “0” |
Examples:
plot_theta.py -m WA1 --skip 50
plot_theta.py -m WA1 -p theta_wa1
Description:
This tool plots the theta distribution resulting from a JAWS stage one simulation.
Two different histograms will be generated. One in which all different copies of the same molecule are added up, and a different one where each copy is displayed individually.
scoop.py¶
Program scoop a protein pdb-file
usage: scoop.py [-h] [-p PROTEIN] [-l LIGAND] [-o OUT] [--center CENTER]
[--innercut INNERCUT] [--outercut OUTERCUT]
[--flexin {sidechain,flexible,rigid}]
[--flexout {sidechain,flexible,rigid}]
[--terminal {keep,doublekeep,neutralize}]
[--excluded EXCLUDED [EXCLUDED ...]]
[--added ADDED [ADDED ...]] [--scooplimit SCOOPLIMIT]
Named Arguments¶
| -p, --protein | the protein PDB-file |
| -l, --ligand | the ligand PDB-file |
| -o, --out | the output PDB-file Default: “scoop.pdb” |
| --center | the center of the scoop, if ligand is not available, either a string or a file with the coordinates Default: “0.0 0.0 0.0” |
| --innercut | maximum distance from ligand defining inner region of the scoop Default: 16.0 |
| --outercut | maximum distance from ligand defining outer region of the scoop Default: 20.0 |
| --flexin | Possible choices: sidechain, flexible, rigid the flexibility of the inner region Default: “flexible” |
| --flexout | Possible choices: sidechain, flexible, rigid the flexibility of the inner region Default: “sidechain” |
| --terminal | Possible choices: keep, doublekeep, neutralize controls of to deal with charged terminal Default: “neutralize” |
| --excluded | a list of indices for residues to be excluded from scoops Default: [] |
| --added | a list of indices for residues to be included in outer scoops Default: [] |
| --scooplimit | the minimum difference between number of residues in protein and scoop for scoop to be retained Default: 10 |
Examples:
scoop.py -p protein.pdb
scoop.py -p protein.pdb -l benzene.pdb
scoop.py -p protein.pdb --center "0.0 0.0 0.0"
scoop.py -p protein.pdb --center origin.dat
scoop.py -p protein.pdb --innercut 10 --outercut 16
scoop.py -p protein.pdb --exclude 189 190
scoop.py -p protein.pdb --added 57 58 59
Description:
This tool truncates a protein and thereby creating a scoop.
All residues outside ocut is removed completely. icut is used to separate the scoop model into two different regions, that possibly can have different sampling regimes. The sampling regimes are determined by --flexin and --flexout.
If the user would like to finetune the residues in the scoop this can be done with --excluded to discard specific residues or --added to include specific residues.
The scoop will be centred on the ligandfile is such a file is provided. Otherwise, it will be centred on the flag --center. The argument to this flag can be either a string with three numbers specifying the centre, as in example three above. It can also be the name of a file containing the centre, as in example four above.
Crystallographic waters that are in proteinfile will also be truncated at ocut
The PDB file will contain specific instructions for ProtoMS to automatically enforce the values of --flexin and --flexout.
solvate.py¶
Program to solvate a solute molecule in either a box or a droplet
usage: solvate.py [-h] [-b BOX] [-s SOLUTE] [-pr PROTEIN] [-o OUT]
[-g {box,droplet,flood}] [-p PADDING] [-r RADIUS]
[-c CENTER] [-n {Amber,ProtoMS}] [--offset OFFSET]
[--setupseed SETUPSEED]
Named Arguments¶
| -b, --box | a PDB-file containing a pre-equilibrated box of water molcules Default: “” |
| -s, --solute | a PDB-file containing the solute molecule |
| -pr, --protein | a PDB-file containing the protein molecule |
| -o, --out | the name of the output PDB-file containing the added water, default solvent_box.pdb Default: “solvent_box.pdb” |
| -g, --geometry | Possible choices: box, droplet, flood the geometry of the added water, should be either ‘box’, ‘droplet’ or ‘flood’ Default: “box” |
| -p, --padding | the minimum distance between the solute and the box edge, default=10 A Default: 10.0 |
| -r, --radius | the radius of the droplet, default=30A Default: 30.0 |
| -c, --center | definition of center, default=’cent’ Default: “cent” |
| -n, --names | Possible choices: Amber, ProtoMS the naming convention, should be either Amber or ProtoMS Default: “ProtoMS” |
| --offset | the offset to be added to vdW radii of the atoms to avoid overfilling cavities with water. Default: 0.89 |
| --setupseed | optional random number seed for generation of water coordinates.. |
if -b or -s are not supplied on the command-line, the program will ask for them.
-c can be either ‘cent’ or a string containing 1, 2 or 3 numbers. If 1 number is given it will be used as center of the droplet in x, y, and z. If 2 numbers are given this is interpreted as an atom range, such that the droplet will be centered on the indicated atoms, and if 3 numbers are given this is directly taken as the center of droplet
- Example usages:
- solvate.py -b ${PROTOMSHOME}/tools/sbox1.pdb -s solute.pdb
- (will solvate ‘solute.pdb’ in a box that extends at least 10 A from
- the solute)
- solvate.py -b ${PROTOMSHOME}/tools/sbox1.pdb -s protein.pdb -g droplet -r 25.0
- (will solvate ‘protein.pdb’ in a 25 A droplet centered on
- all coordinates)
Examples:
solvate.py -b $PROTOMSHOME/data/wbox_tip4p.pdb -s benzene.pdb
solvate.py -b $PROTOMSHOME/data/wbox_tip4p.pdb -s benzene.pdb -p 12.0
solvate.py -b $PROTOMSHOME/data/wbox_tip4p.pdb -s benzene.pdb -pr protein.pdb -g droplet
solvate.py -b $PROTOMSHOME/data/wbox_tip4p.pdb -s benzene.pdb -pr protein.pdb -g droplet -r 24.0
solvate.py -b $PROTOMSHOME/data/wbox_tip4p.pdb -pr protein.pdb -g droplet -c 0.0
solvate.py -b $PROTOMSHOME/data/wbox_tip4p.pdb -pr protein.pdb -g droplet -c "0.0 10.0 20.0"
solvate.py -b $PROTOMSHOME/data/wbox_tip4p.pdb -pr protein.pdb -g droplet -c "76 86"
solvate.py -b $PROTOMSHOME/data/wbox_tip4p.pdb -s gcmc_box.pdb -g flood
Description:
This tool solvates a ligand in either a droplet or a box of water. It can also flood a GCMC or JAWS-1 simulations box with waters.
Pre-equilibrated boxes to use can be found in the $PROTOMSHOME/data folder.
To solvate small molecule it is sufficient to give the solutefile as in the first example above. This produces a box with at least 10 A between the solute and the edge of the water box, which should be sufficient in most situation. Use padding to increase or decrease the box size as in the second example. The solvation box is created by replicating the pre-equilibrated box in all dimensions and then removing waters that overlap with solute atoms.
To solvate a protein in a droplet, specify proteinfile and droplet as in the third example above. This produces a droplet with radius of 30 A, which was chosen to work well with the default options in scoop.py. Use radius to obtain a smaller or larger droplet as in the fourth example. The centre of the droplet can be on a ligand if ligandfile is specified. Otherwise, the center``argument is used. This argument can be either ``cent (the default) that places the droplet at the centre of the protein. It can also take a single number as in the fifth example above in case it is placed at this coordinate in all dimensions. It can also take a string with three numbers which is the origin of the droplet in x, y, and z dimensions, see the sixth example above. If two numbers are given as in the seventh example above, it is assumed that this is an atom range and the droplet will be placed at the centre of these atoms. The droplet is created by putting random waters from the pre-equilibrated box on a grid, displacing them slightly in a random fashion.
The tool can also be used to fill a box with waters for GCMC and JAWS-1 simulations, similar to distribute_waters.py. In this case the solute is typically a box created by make_gcmcbox.py and flood needs to be specified, see the last example above. This gives a box filled with the bulk number of waters.
split_jawswater.py¶
Program to split JAWS-1 waters to a number of PDB-files for JAWS-2
usage: split_jawswater.py [-h] [-w WATERS] [-o OUT] [--jaws2box]
Named Arguments¶
| -w, --waters | the name of the PDB-file containing the waters. |
| -o, --out | the prefix of the output PDB-files Default: “” |
| --jaws2box | whether to apply a header box for jaws2 to the pdb files of individual waters Default: False |
Examples:
split_jawswater.py -w waters.pdb
split_jawswater.py -w waters.pdb -o jaws2_
Description:
This tool splits a PDB file containing multiple water molecules into PDB files appropriate for JAWS-2.
For each water molecule in pdbfile the tool will write a PDB file with individual water molecules named outprefix+watN.pdb where N is the serial number of the water molecule. Furthermore, the tool will write a PDB file with all the other molecules and name if outprefix+notN.pdb where again N is the serial number of the water molecule. In these latter PDB-files, the water residue name is changed to that of the bulk water, e.g., t3p or t4p.
For instance, if waters.pdb in the second example above contains 3 water molecule, this tool will create the following files:
jaws2_wat1.pdb
jaws2_wat2.pdb
jaws2_wat3.pdb
jaws2_not1.pdb
jaws2_not2.pdb
jaws2_not3.pdb
