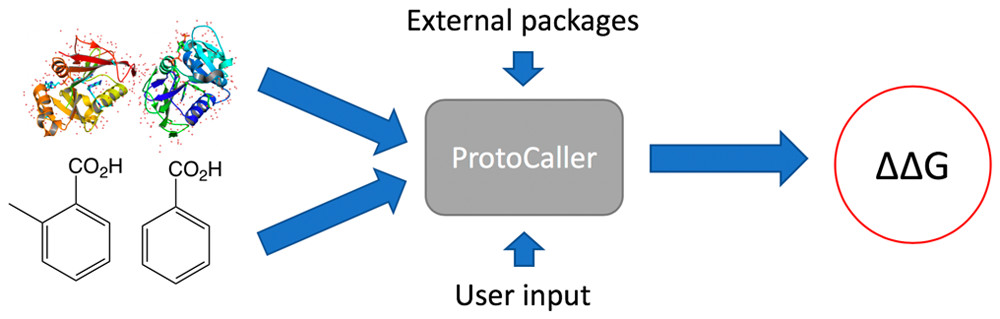
I am a final year PhD student, part of the TMCS (Theory and Modelling in Chemical Sciences) CDT and our research focuses on alchemical protein-ligand binding free energy calculations using classical molecular mechanics models. We collaborate with AstraZeneca, GSK, Syngenta and Sygnature Discovery to investigate the reproducibility, repeatability and general robustness of these calculations, in order to increase their impact in the pharmaceutical industry.I have so far developed a Python library which aids the high-throughput preparation and execution of binding free energy calculations (ProtoCaller: paper, github) and is also a part of a general online platform (GalaxyTools: paper, github, in collaboration with the University of Cape Town). I have afterwards used ProtoCaller to conduct a large-scale sensitivity study of binding free energy calculations to their initial conditions (not yet published). My current project concentrates on developing a robust enhanced sampling method to increase the reliability and performance of alchemical free energy calculations.