The group's github page can be found at https://github.com/essex-lab/. Below is a list of the software that has been developed and published by the group.
pip install openmmslicer. OpenMM and OpenMMTools are required.
conda install paramol -c jmorado -c ambermd -c conda-forge -c omnia -c rdkit -c anaconda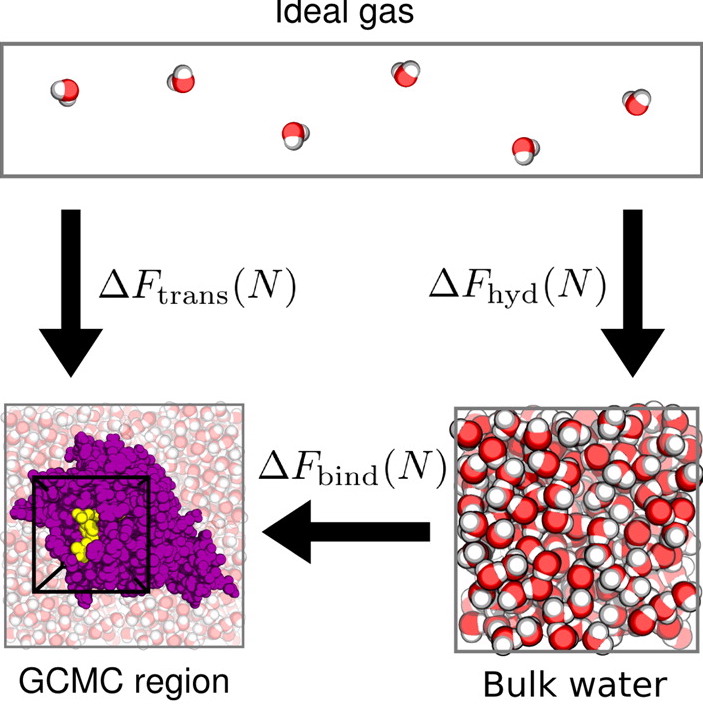
conda install -c omnia -c anaconda -c conda-forge -c essexlab grand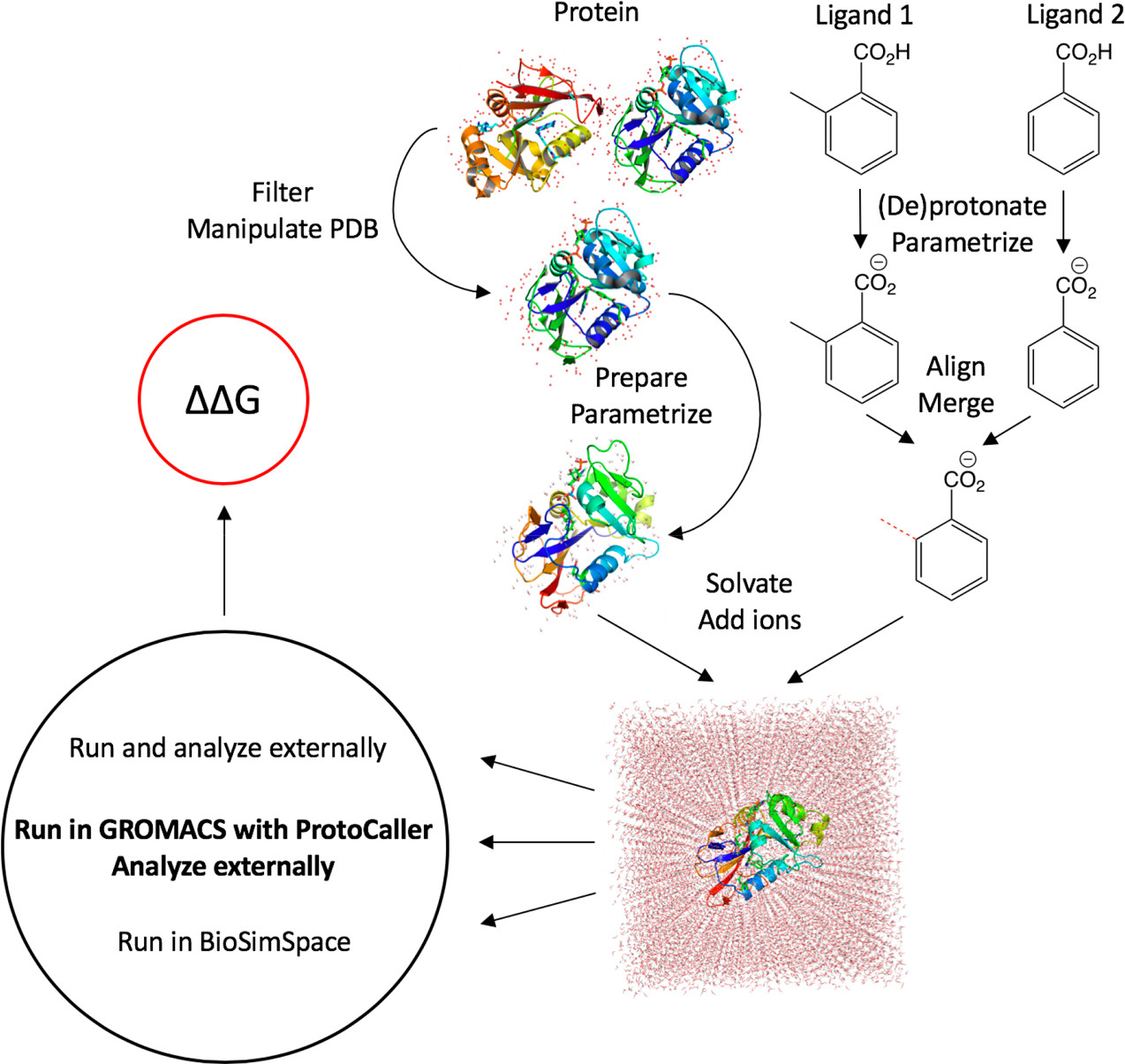
conda install -c conda-forge -c omnia -c michellab -c essexlab protocaller